Webinar: Why Spatial Proteomics Matters for Understanding the Tumor Microenvironment
A Dana-Farber Case Study — Hodgkin Lymphoma
The webinar, “High-Definition Spatial Proteomics to Elucidate the Tumor Immune Microenvironment: A Dana-Farber Case Study” was presented by pathologists Scott Rodig and Kyle Wright, along with Ionpath scientist Yari Sigal. It’s now available to view on-demand. But if you don’t have the time to watch, we’ve summed up some highlights here.
The case study focused on classical Hodgkin lymphoma, a type of cancer in which malignant cells can be quite rare in patient biopsies — as little as 1% of the total cellular infiltrate in the mass that’s biopsied. Rodig, who kicked off the presentation with a quick introduction to this type of cancer and how it evades immune response, noted that these cells are characterized by a high tumor mutation burden, and as many as 30% of them also carry Epstein-Barr virus.
These traits illustrate why spatial analysis is so important. Developing immunotherapies to treat this type of Hodgkin lymphoma will require a significantly improved understanding of immunological biomarkers and the individual cell phenotypes present in the tumor microenvironment as well as immune evasion tactics used by the cancer. While standard chemotherapy has been effective for most patients, the cure rate drops to just 20% for cases with recurrent or refractory cancer. Immunotherapies appear promising for classical Hodgkin lymphoma, which often deploys the same expression of PD-1 ligands seen in other cancers that have been effectively targeted with newer immuno-oncology treatments. Knowledge of the tumor microenvironment will be essential for making progress.
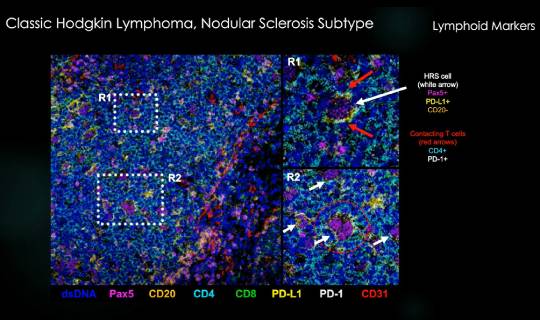
Key cellular insights enabled by MIBI
During Sigal’s part of the webinar, he introduced viewers to MIBI technology, which was designed to work within a pathologist’s existing workflow. Our high-resolution imaging tool using time-of-flight mass spectrometry paired with our MIBItracker data visualization and analysis suite, allows scientists to interrogate and quantify up to 40 markers from an FFPE sample in a process that preserves sample for future use. The highly reproducible process is able to detect the full biological range of proteins in a sample, from single antibodies to the most abundant proteins. MIBI data can be used for high-definition single cell analysis, comprehensive cell type enumeration and quantitative expression analysis in spatial context .
The audience Q&A portion of the webinar covered great technical detail, such as analysis software packages with which MIBI is compatible, sample types beyond FFPE that can be studied, imaging time, and more.
Finally, Rodig noted that based on the useful data generated by MIBI for the Hodgkin study, DFCI scientists have expanded their use of this technology to other cancers. Going forward, they will also be using it to produce much-needed spatial proteomic data about a broad range of cancer types for the Human Tumor Atlas project.
Interested in learning more? Request a pilot project or check out details about MIBI technology.