To Parse Neurological Disorders, Scientists Map Protein Markers with MIBI™ Technology
The preprint, recently posted to bioRxiv, comes from lead authors Kausalia Vijayaragavan and Bryan Cannon, senior author Sean Bendall, and collaborators. In it, the researchers describe using multiplex ion beam imaging (MIBI™) technology to study aberrant protein expression and accumulation in archived brain samples from individuals with cognitive impairment, Alzheimer’s disease, and other conditions.
The project was conceived to overcome limitations of conventional tools to characterize brain tissue, which is typically challenging to analyze using fluorescence-based techniques. “Our understanding of the underlying phenotypic, regional, and cellular heterogeneity in neurodegenerative niches of these vulnerable brain regions remains limited,” the authors note.
Turning to MIBI spatial proteomic technology
Because MIBI technology is based on mass spectrometry and elemental mass tags, it is not confounded by the challenges of endogenous autofluorescence from tissue –especially matrix-fixed tissue–that hinder fluorescence-based tools. That made it an ideal candidate for analyzing formalin-fixed, paraffin-embedded (FFPE) brain samples. The resolution of MIBI technology was another key advantage: its nanometer scale means it can deliver results at the level of individual cells.
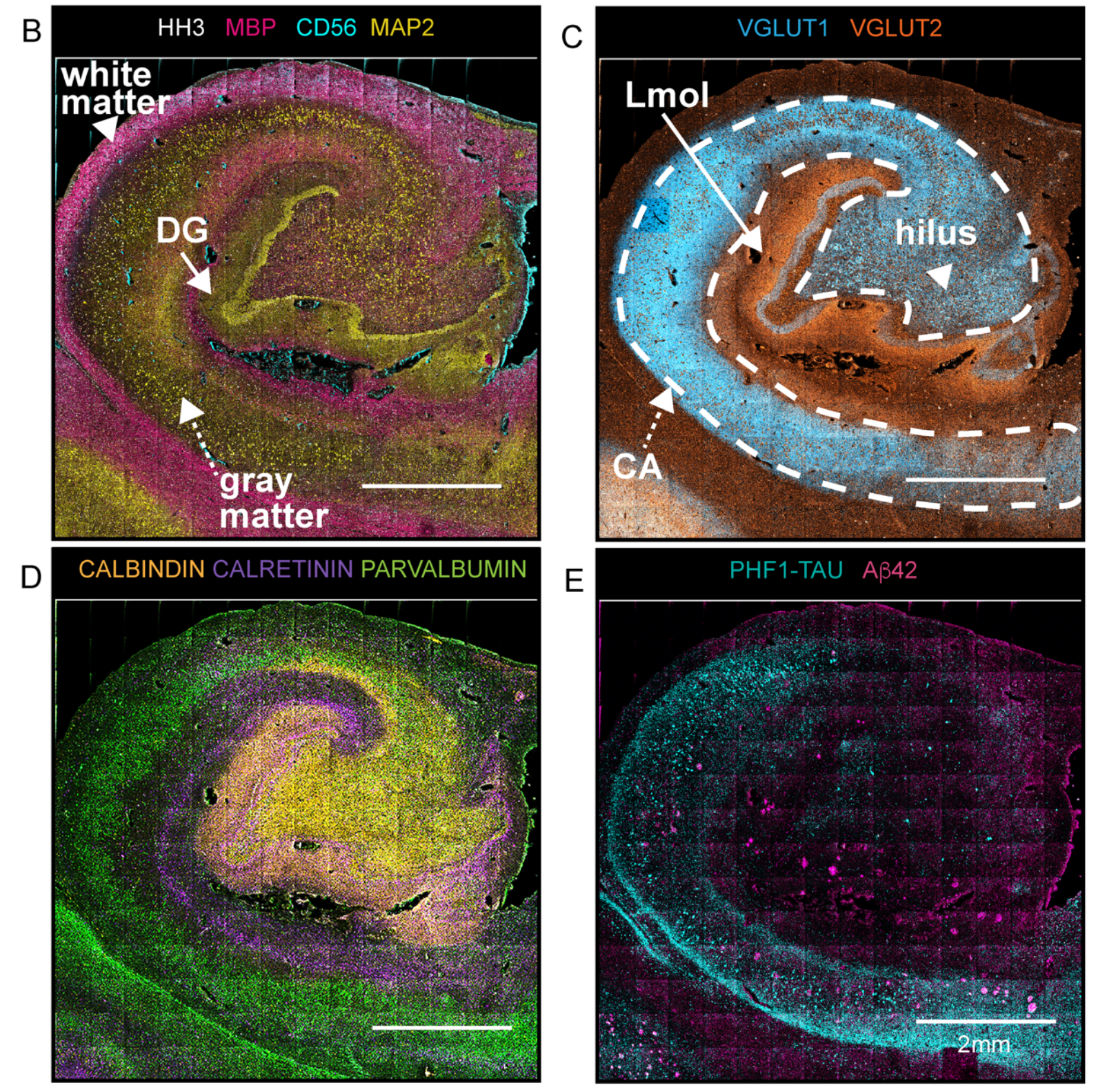
To demonstrate this approach, the team validated antibodies for specific brain protein targets and combined them in a 36-antibody panel. “To capture salient neuropathologic changes, the antibody staining panel targeted proteins for delineating major neuronal cell lineages, proteopathies, and vascular structures,” the authors report.
With that panel in hand, the scientists chose archived FFPE samples from three individuals, along with carefully selected controls. They deployed MIBI technology and the 36-antibody panel to resolve targeted proteins across the hippocampus.

Analyses underscore the importance of proteomic resolution
This approach led to impressive detail in the team’s analyses. “We extracted and classified a wide range of brain cellular features and protein aggregates,” they report. “Our imaging panel was designed to phenotype brain cellular, structural, and proteopathy features while displaying well-known brain topologies.”
In the brain sample affected by Alzheimer’s disease, for example, scientists used nuclear segmentation to identify more than 15,000 cells; a pixel-based technique identified nearly 20,000 features that were not connected to cell nuclei, such as amyloid-β peptide plaques. Vijayaragavan et al. performed a number of analyses to characterize proteomic differences between healthy brains and those with dementia.
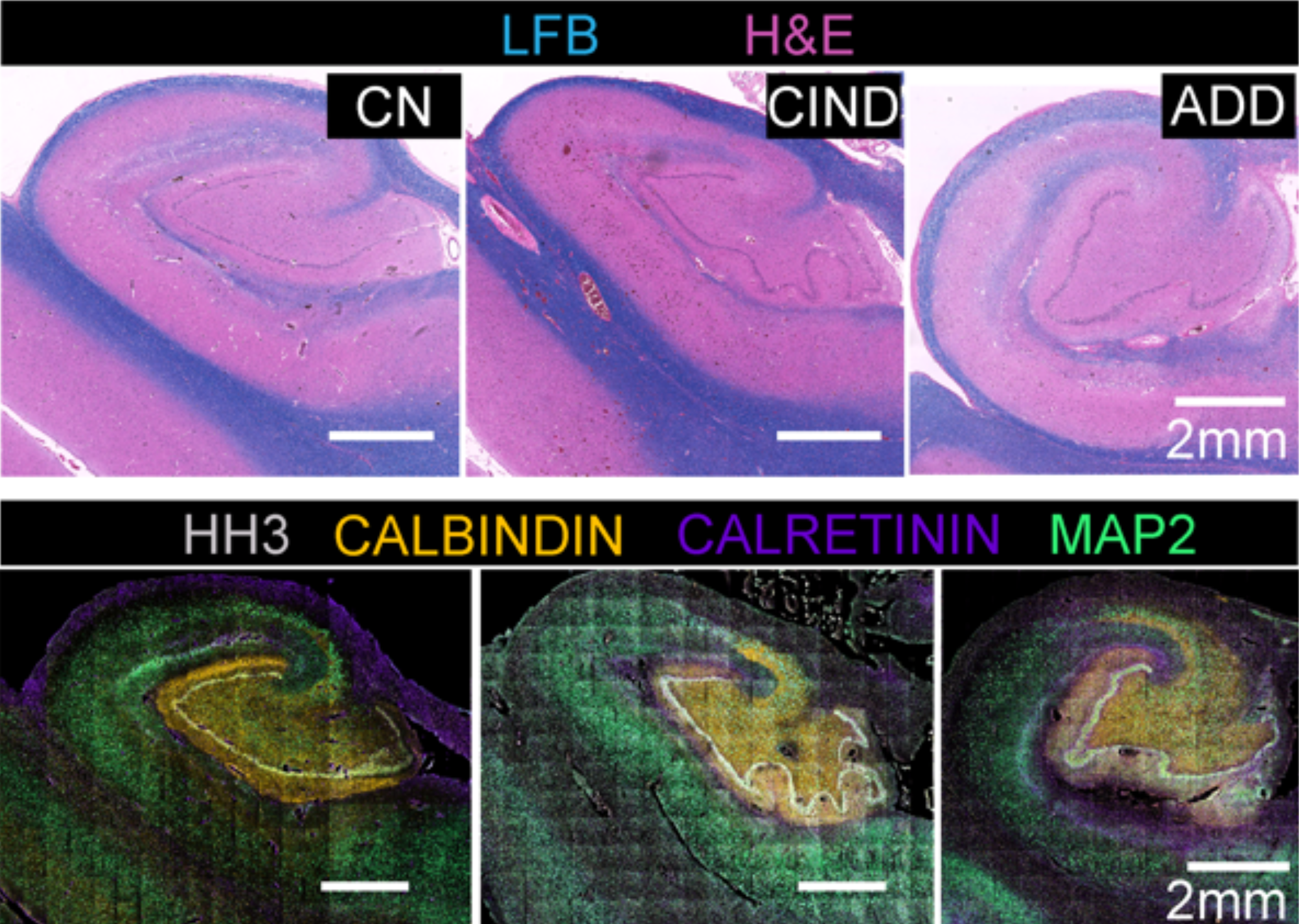
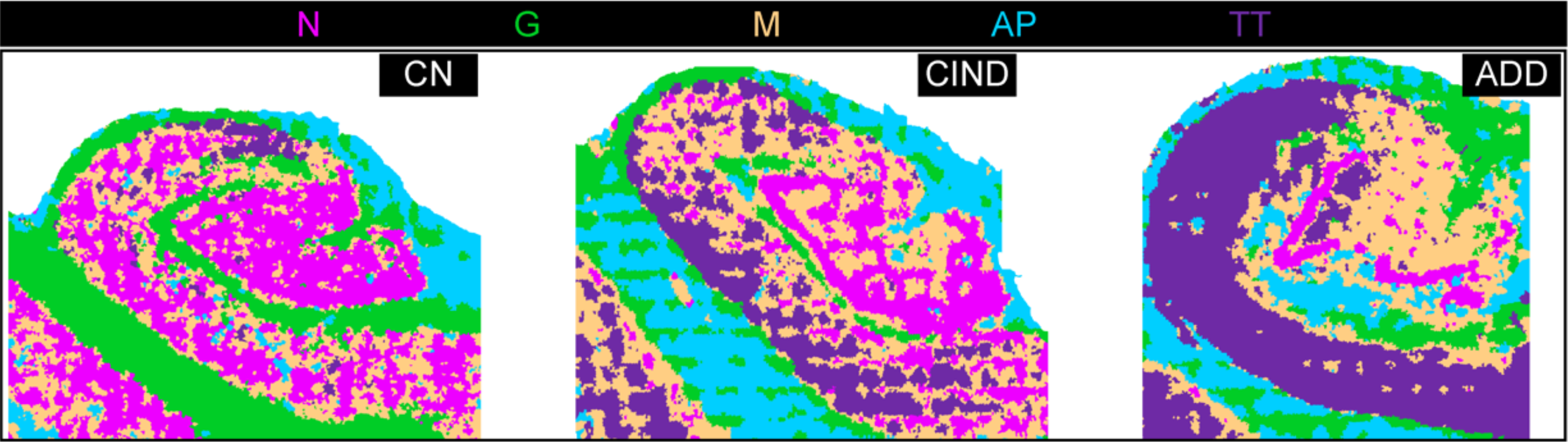
The team also used this approach to analyze a tissue microarray of six brain regions from individuals with no known neurological disorders and those with some cognitive impairment. “Combinatorial protein expression patterns provided a snapshot of functional organization or proteopathy within brain regions,” the scientists write. “This low-level analysis of multiplexed proteomic data could serve as a guide for ‘fingerprinting’ human brain and be used to model progression in neurodegeneration.”
The scientists believe this project could inform subsequent studies of neurological disorders based on MIBI technology and their analytical pipeline. “Our study revealed unique insights from multiplexed imaging and data-driven approaches for neuropathologic analysis and serves as a baseline for mechanistic and interventional understanding in human neurodegeneration,” they write. “Integration of this spatially multiplexed data with further clinical histories will drive insights on possible cellular mechanisms underlying symptomatic disease progression or resilience in [central nervous system] disease.”