Scientists Deploy MIBI Technology for Spatial Analysis of HIV Infection Model
New approach provides spatial multi-omic analysis of an HIV model
In a new preprint posted to bioRxiv, scientists at Stanford University, Oregon Health & Science University, and the Dana-Farber Cancer Institute report findings from the multi-omic analysis of an HIV model. The study was used to validate a new approach they call PANINI (short for protein and nucleic acid in situ imaging) that can be paired with multiplexed ion beam imaging (MIBI™) technology to study proteins and nucleic acids in the same tissue sample.
Eradication of HIV will require a deeper understanding of how the virus hunkers down in host cells, creating tissue reservoirs that are remarkably difficult for the body’s immune system to find and fight off. “To understand the mechanisms and pathology of HIV persistence it is necessary to visualize the tissue microenvironments where the virus resides,” writes Sizun Jiang, one of the lead authors on the manuscript. “However, technological barriers have limited our ability to phenotypically characterize and quantify the cellular components of viral tissue reservoirs.”
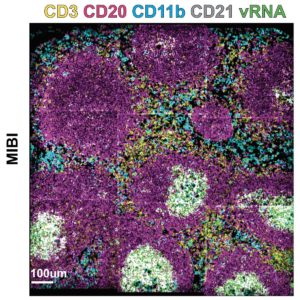
Spatial biology platforms have empowered visualization of biomolecules, but what the scientists really wanted was a comprehensive view of proteins, DNA, and RNA at the same time — not just one type of analyte. Conventional ISH approaches typically require a processing step that is not ideal with simultaneous protein detection, so a new approach was needed.
PANINI-MIBI enables RNA, DNA, and protein imaging in FFPE tissue
The team turned to a new method that they named PANINI. Designed to be paired with a system like our MIBI technology, this approach carefully preserves both nucleic acids and proteins by circumventing protease digestion for a spatially resolved, multi-omic view of the tissue sample.
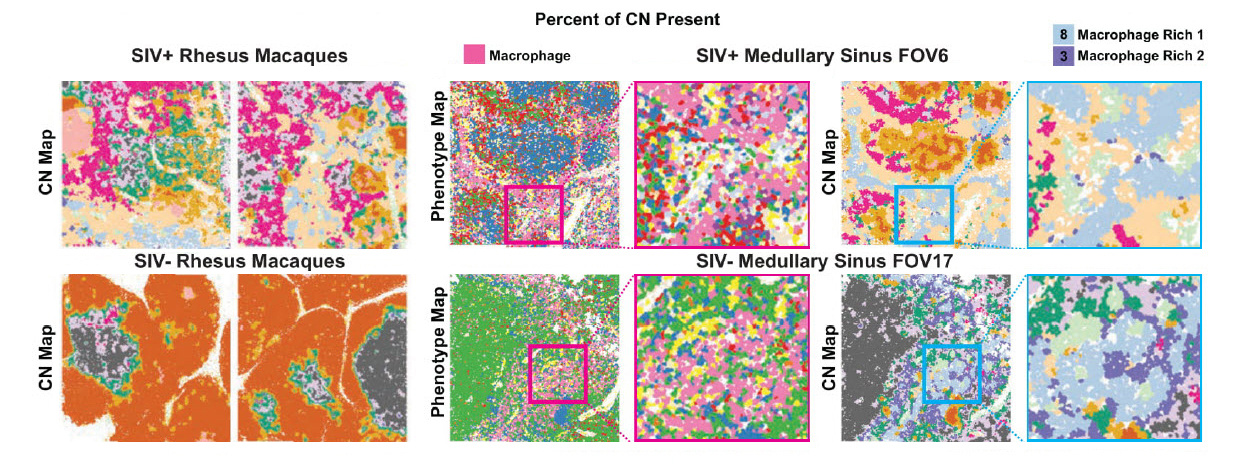
To demonstrate its utility, scientists used the PANINI-MIBI combination to analyze more than 30 parameters in FFPE samples and lymphoid tissues from rhesus macaques, studying both healthy animals and ones infected with simian immunodeficiency virus (SIV), a model for HIV. This allowed them “to characterize in unprecedented detail the viral reservoir and immune responses,” the team reports. “This enabled multiplexed dissection of cellular phenotypes, functional markers, viral DNA integration events, and viral RNA transcripts as resulting from viral infection.” They identified 11 types of cellular neighborhoods and found markers that could differentiate among healthy cells, cells with acute infection, and cells with chronic infection.
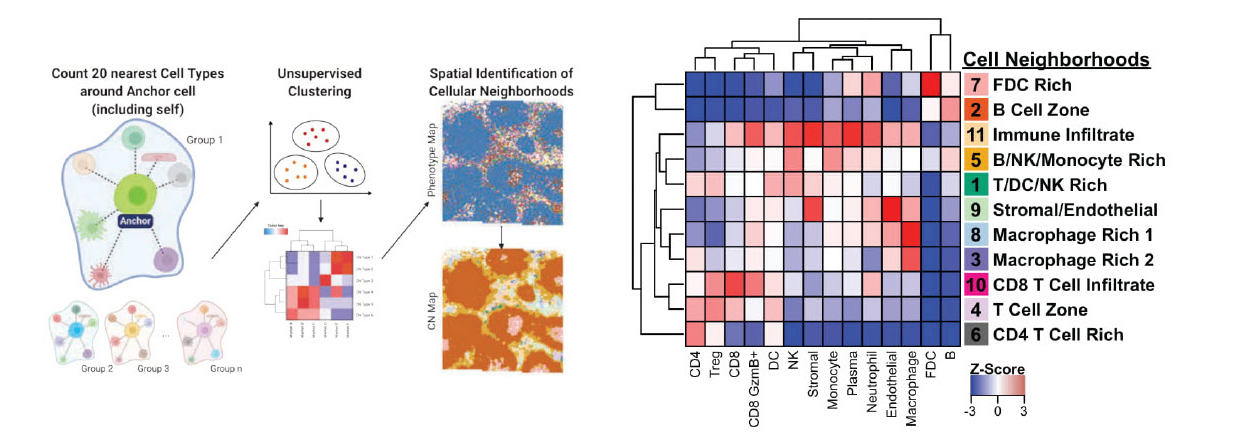
Unlocking the mystery of cellular reservoirs in HIV
The researchers identified a number of traits that appear correlated with virus reservoirs. For instance, infected cells were more likely to be found in regions with dense B cell populations. They tend not to transcribe viral genes when they’re near the vascular system, but often produce transcripts when they’re near follicular dendritic cells. “Additional contributing features that distinguish between viral latency and transcription include the expression of CD56, and quantities of Tregs, neutrophils and CD8 T cells,” they add.

This study was lead by postdoctoral fellows Sizun Jiang, Ph.D (Stanford), Chan Chi Ngan (OHSU) and Xavier Rovira-Clave (Stanford), with senior authors Michael Angelo (Stanford), Jacob Estes (OHSU) and Garry Nolan (Stanford). The study was generously supported by various funding agencies, including the National Institutes of Health, the US Food and Drug Administration Medical Countermeasures Initiative and the Bill and Melinda Gates Foundation.