Featured Publications
Researchers used spatial proteomics to analyze a model of HIV, validating a new approach that allows for the study of proteins and nucleic acids in the same tissue sample
An approach that combines protein and nucleic acid imaging reveals virus-dependent B cell and macrophage immunosuppression of tissue microenvironments
Jiang, et al., Immunity 20 April 2022
Key Insights
- Scientists developed PANINI (short for protein and nucleic acid in situ imaging), which can be paired with MIBI™ spatial proteomics technology. The approach skips protease digestion to preserve nucleic acids and proteins for comprehensive analysis.
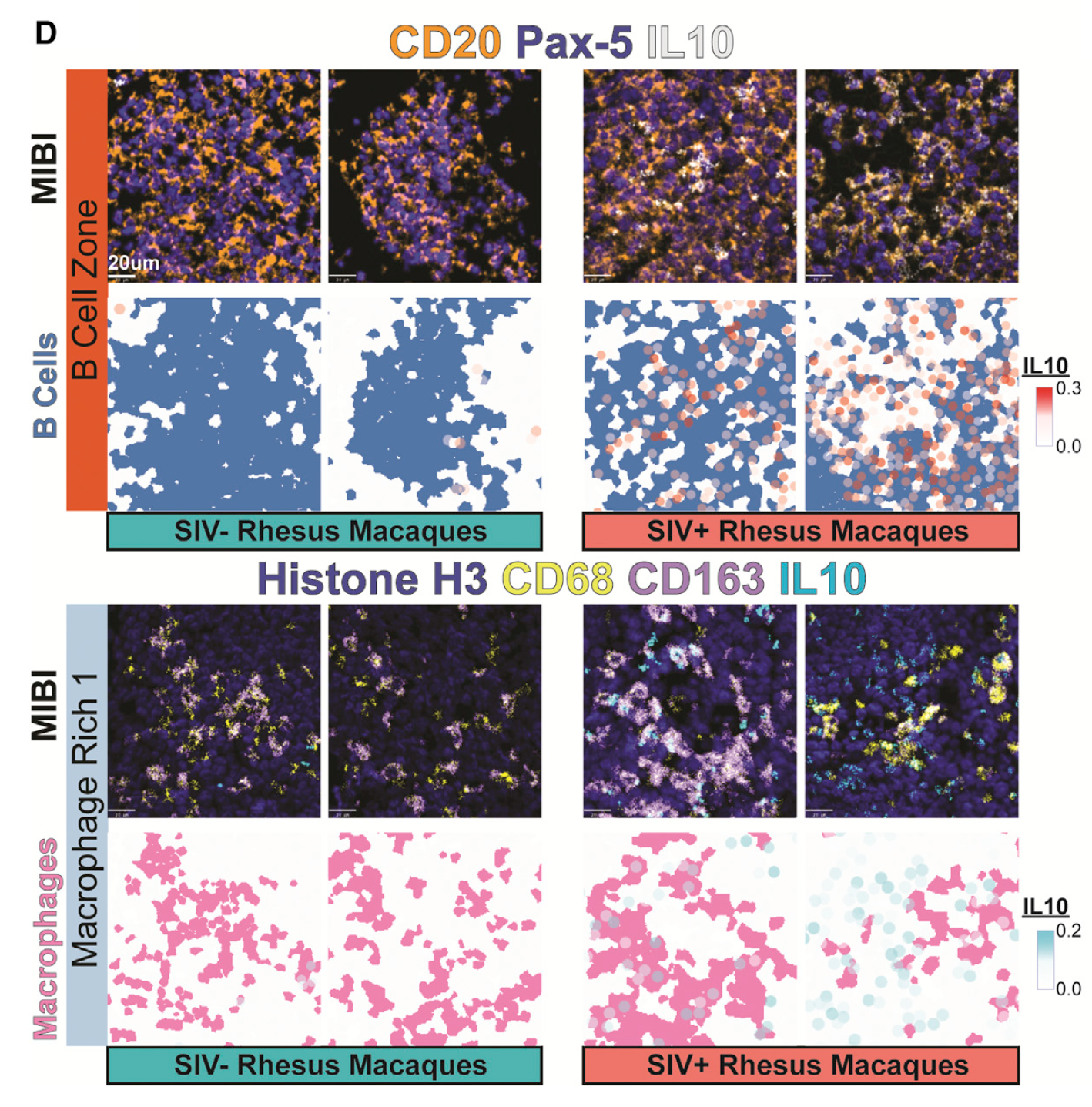
- They used the PANINI-MIBI combination to analyze more than 30 parameters in FFPE samples and lymphoid tissues from rhesus macaques, detecting 11 types of cellular neighborhoods and identifying markers that could differentiate among healthy cells, cells with acute infection, and cells with chronic infection.
- Along the way, the scientists found several traits that appear to be correlated with virus reservoirs, such as regions with dense B cell populations or proximity to follicular dendritic cells.
Abstract
Understanding the mechanisms of HIV tissue persistence necessitates the ability to visualize tissue microenvironments where infected cells reside; however, technological barriers limit our ability to dissect the cellular components of these HIV reservoirs. Here, we developed protein and nucleic acid in situ imaging (PANINI) to simultaneously quantify DNA, RNA, and protein levels within these tissue compartments. By coupling PANINI with multiplexed ion beam imaging (MIBI), we measured over 30 parameters simultaneously across archival lymphoid tissues from healthy or simian immunodeficiency virus (SIV)-infected nonhuman primates. PANINI enabled the spatial dissection of cellular phenotypes, functional markers, and viral events resulting from infection. SIV infection induced IL-10 expression in lymphoid B cells, which correlated with local macrophage M2 polarization. This highlights a potential viral mechanism for conditioning an immunosuppressive tissue environment for virion production. The spatial multimodal framework here can be extended to decipher tissue responses in other infectious diseases and tumor biology.