Recent Publications
Stanford team uses Ionpath spatial proteomics in multimodal approach and reveals tumor-specific keratinocyte population at leading edges of squamous cell carcinomas
Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma
Ji, et al., Cell 23 Jul 2020: (182), 1-18.
Key Insights
- A new tumor cell subpopulation of tumor-specific keratinocytes (TSKs) was identified and observed to play a key role in intercellular communication
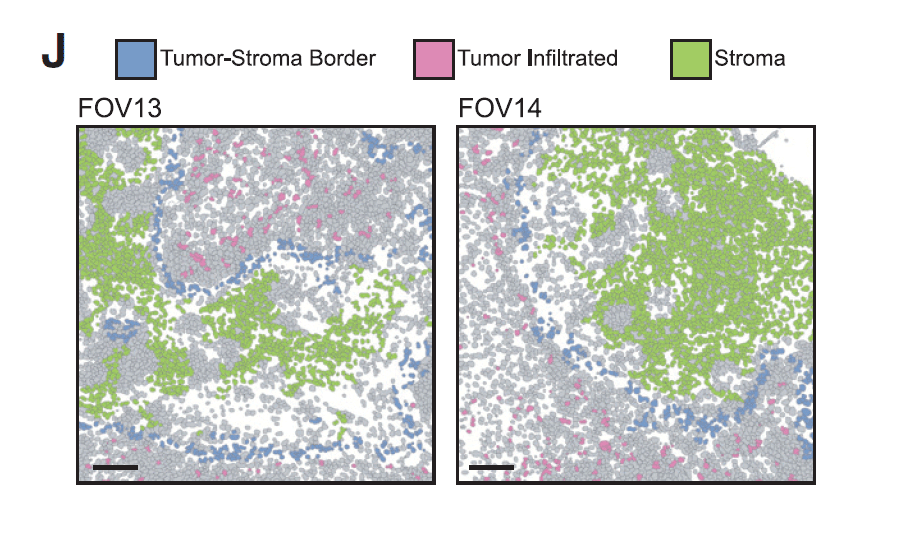
- TSKs were found to reside at the leading edges of carcinoma tissue in fibrovascular niches
- Multiplexed ion beam imaging (MIBI) enabled spatial mapping of tumor, stromal, and immune cells with true single-cell resolution and revealed key intercellular spatial relationships
- A multimodal approach for investigating tumor microenvironments enabled a more comprehensive understanding of tumor biology
Abstract
To define the cellular composition and architecture of cutaneous squamous cell carcinoma (cSCC), Ji et al. combined single-cell RNA sequencing with spatial transcriptomics and multiplexed ion beam imaging (MIBI, a multiplexed spatial proteomics platform) from a series of human cSCCs and matched normal skin. cSCC exhibited four tumor subpopulations, three recapitulating normal epidermal states, and a tumor-specific keratinocyte (TSK) population unique to cancer, which localized to a fibrovascular niche. Integration of single-cell and spatial data mapped ligand-receptor networks to specific cell types, revealing TSK cells as a hub for intercellular communication. Multiple features of potential immunosuppression were observed, including T regulatory cell (Treg) co-localization with CD8 T cells in compartmentalized tumor stroma. Finally, single-cell characterization of human tumor xenografts and in vivo CRISPR screens identified essential roles for specific tumor subpopulation-enriched gene networks in tumorigenesis. These data define cSCC tumor and stromal cell subpopulations, the spatial niches where they interact, and the communicating gene networks that they engage in cancer.
View the data on our interactive MIBItracker demo site >