Publications & Posters
Automated Single Cell Phenotyping of Time-of-Flight Secondary Ion Mass Spectrometry Tissue Images<span class='publication-meta'>Bajaj, <i>et al.</i>, <i>J. Am. Soc. Mass Spectrom.</i> November 2024</span>
Existing analytical techniques are being improved or applied in new ways to profile the tissue microenvironment (TME) to better understand the role of cells in disease research. Fully understanding the complex interactions between cells of many different types and functions is often slowed by the intense data analysis required. Multiplexed Ion Beam Imaging (MIBI) has been developed to simultaneously characterize 50+ cell types and their functions within the TME with a subcellular spatial resolution, but this results in complex data sets that are challenging to qualitatively analyze. Deep Learning (DL) techniques were used to build the MIBIsight workflow, which can process images containing thousands of cells into easily digestible reports and plots to enable researchers to easily summarize data sets in a study and make informed conclusions. Here we present the three types of DL models that have been trained with annotated MIBI images that have been pathologist validated as well as the associated workflow for the evolution of raw mass spectral data into actionable reports and plots.
Bajaj, et al., J. Am. Soc. Mass Spectrom. November 2024
doi: https://doi.org/10.1021/jasms.4c00328
Highly Multiplexed Tissue Imaging in Precision Oncology and Translational Cancer Research<span class='publication-meta'>Bollhagen, <i>et al.</i>, <i>Cancer Discov.</i> November 2024</span>
Precision oncology tailors treatment strategies to a patient's molecular and health data. Despite the essential clinical value of current diagnostic methods, hematoxylin and eosin morphology, immunohistochemistry, and gene panel sequencing offer an incomplete characterization. In contrast, highly multiplexed tissue imaging allows spatial analysis of dozens of markers at single-cell resolution enabling analysis of complex tumor ecosystems; thereby it has the potential to advance our understanding of cancer biology and supports drug development, biomarker discovery, and patient stratification. We describe available highly multiplexed imaging modalities, discuss their advantages and disadvantages for clinical use, and potential paths to implement these into clinical practice. Significance: This review provides guidance on how high-resolution, multiplexed tissue imaging of patient samples can be integrated into clinical workflows. It systematically compares existing and emerging technologies and outlines potential applications in the field of precision oncology, thereby bridging the ever-evolving landscape of cancer research with practical implementation possibilities of highly multiplexed tissue imaging into routine clinical practice.
Bollhagen, et al., Cancer Discov. November 2024
doi: https://doi.org/10.1158/2159-8290.CD-23-1165
Spatial analysis by current multiplexed imaging technologies for the molecular characterisation of cancer tissues<span class='publication-meta'>Semba, <i>et al.</i>, <i>Br J Cancer</i> October 2024</span>
Abstract:
Tumours are composed of tumour cells and the surrounding tumour microenvironment (TME), and the molecular characterisation of the various elements of the TME and their interactions is essential for elucidating the mechanisms of tumour progression and developing better therapeutic strategies. Multiplex imaging is a technique that can quantify the expression of multiple protein markers on the same tissue section while maintaining spatial positioning, and this method has been rapidly developed in cancer research in recent years. Many multiplex imaging technologies and spatial analysis methods are emerging, and the elucidation of their principles and features is essential. In this review, we provide an overview of the latest multiplex imaging techniques by type of imaging and staining method and an introduction to image analysis methods, primarily focusing on spatial cellular properties, providing deeper insight into tumour organisation and spatial molecular biology in the TME.
Semba, et al., Br J Cancer October 2024
doi: https://doi.org/10.1038/s41416-024-02882-6
Myeloid-instructed CD14+CD4+ T cells within the tumor microenvironment are associated with TNF⍺ signaling and prolonged survival in non-small cell lung cancer<span class='publication-meta'>Marceaux, <i>et al.</i>, <i>bioRxiv</i> August 2024</span>
Abstract:
Immune cells from the adaptive and innate immune systems coexist in the tumor immune microenvironment (TME) to mediate tumor cell immunosurveillance and prevent tumor relapse. Relapse-free survival after surgical resection is heterogeneous in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), the two major subtypes of non-small cell lung cancer. Yet, little is known about the composition and spatial organisation of the TME that could explain the different prognoses for lung cancer patients. Using spatial multi-omics analyses, we show that LUAD and LUSC have distinct TMEs and that neutrophil-enriched tumors are associated with worse survival, while the accumulation of myeloid-instructed CD14+ T cells in the tumor core was associated with prolonged patient survival. CD14+CD4+ T cells express prototypic markers of myeloid cells and were found to be clonally expanded tumor-specific activated T cells. TNFα signaling was activated in tumors with high infiltration of CD14+CD4+ T cells in the tumor core. Our results demonstrate that innate immune cells impact adaptive immune cell activity to support the deployment of discrete T cell populations with anti-tumor activity.
Marceaux, et al., bioRxiv August 2024
doi: https://doi.org/10.1101/2024.08.02.606259
High-plex imaging and cellular neighborhood spatial analysis reveals multiple immune escape and suppression patterns in diffuse large B-cell lymphoma<span class='publication-meta'>Reiss, <i>et al.</i>, <i>Leukemia</i> May 2024</span>
A hallmark of hematological malignancies such as diffuse large B-cell lymphoma (DLBCL) is heterogeneity, with contribution from the tumor and its microenvironment. Application of next generation sequencing technology to patient tumor biopsies revealed not only genetic and epigenetic underpinnings of tumor intrinsic heterogeneity [1, 2], but also the complexities of the tumor microenvironment (TME). Furthermore, the lymphoma microenvironment has been characterized through computational inference or digital cytometry techniques from large collections of DLBCL gene expression data [3, 4]. These analyses demonstrated that despite lacking a clear structure of tumor/immune compartments found in solid tumors, the lymphoma TME is not a random assortment of tumor and immune cells. We believe the spatial relationship between tumor and infiltrating immune cells is a missing piece in our understanding.
Reiss, et al., Leukemia May 2024
doi: https://doi.org/10.1038/s41375-024-02239-1
Phase 1 dose expansion and biomarker study assessing first-in-class tumor microenvironment modulator VT1021 in patients with advanced solid tumors<span class='publication-meta'>Chen, <i>et al.</i>, <i>Commun Med (Lond)</i> March 2024</span>
Background: Preclinical studies have demonstrated that VT1021, a first-in-class therapeutic agent, inhibits tumor growth via stimulation of thrombospondin-1 (TSP-1) and reprograms the tumor microenvironment. We recently reported data from the dose escalation part of a phase I study of VT1021 in solid tumors. Here, we report findings from the dose expansion phase of the same study.
Methods: We analyzed the safety and tolerability, clinical response, and biomarker profile of VT1021 in the expansion portion of the phase I study (NCT03364400). Safety/tolerability is determined by adverse events related to the treatment. Clinical response is determined by RECIST v1.1 and iRECIST. Biomarkers are measured by multiplexed ion beam imaging and enzyme-linked immunoassay (ELISA).
Results: First, we report the safety and tolerability data as the primary outcome of this study. Adverse events (AE) suspected to be related to the study treatment (RTEAEs) are mostly grade 1-2. There are no grade 4 or 5 adverse events. VT1021 is safe and well tolerated in patients with solid tumors in this study. We report clinical responses as a secondary efficacy outcome. VT1021 demonstrates promising single-agent clinical activity in recurrent GBM (rGBM) in this study. Among 22 patients with rGBM, the overall disease control rate (DCR) is 45% (95% confidence interval, 0.24-0.67). Finally, we report the exploratory outcomes of this study. We show the clinical confirmation of TSP-1 induction and TME remodeling by VT1021. Our biomarker analysis identifies several plasmatic cytokines as potential biomarkers for future clinical studies.
Conclusions: VT1021 is safe and well-tolerated in patients with solid tumors in a phase I expansion study. VT1021 has advanced to a phase II/III clinical study in glioblastoma (NCT03970447).
Chen, et al., Commun Med (Lond) March 2024
doi: https://doi.org/10.1038/s43856-024-00520-z
A Hitchhiker’s guide to high-dimensional tissue imaging with multiplexed ion beam imaging<span class='publication-meta'>Yeo, <i>et al.</i>, <i>Methods Cell Biol.</i> March 2024</span>
Advancements in multiplexed tissue imaging technologies are vital in shaping our understanding of tissue microenvironmental influences in disease contexts. These technologies now allow us to relate the phenotype of individual cells to their higher-order roles in tissue organization and function. Multiplexed Ion Beam Imaging (MIBI) is one of such technologies, which uses metal isotope-labeled antibodies and secondary ion mass spectrometry (SIMS) to image more than 40 protein markers simultaneously within a single tissue section. Here, we describe an optimized MIBI workflow for high-plex analysis of Formalin-Fixed Paraffin-Embedded (FFPE) tissues following antigen retrieval, metal isotope-conjugated antibody staining, imaging using the MIBI instrument, and subsequent data processing and analysis. While this workflow is focused on imaging human FFPE samples using the MIBI, this workflow can be easily extended to model systems, biological questions, and multiplexed imaging modalities.
Yeo, et al., Methods Cell Biol. March 2024
doi: https://doi.org/10.1016/bs.mcb.2024.02.018
A Spatial Multi-Modal Dissection of Host-Microbiome Interactions within the Colitis Tissue Microenvironment<span class='publication-meta'>Zhu, <i>et al.</i>, <i>bioRxiv</i> March 2024</span>
Abstract:
The intricate and dynamic interactions between the host immune system and its microbiome constituents undergo dynamic shifts in response to perturbations to the intestinal tissue environment. Our ability to study these events on the systems level is significantly limited by in situ approaches capable of generating simultaneous insights from both host and microbial communities. Here, we introduce Microbiome Cartography (MicroCart), a framework for simultaneous in situ probing of host features and its microbiome across multiple spatial modalities. We demonstrate MicroCart by comprehensively investigating the alterations in both gut host and microbiome components in a murine model of colitis by coupling MicroCart with spatial proteomics, transcriptomics, and glycomics platforms. Our findings reveal a global but systematic transformation in tissue immune responses, encompassing tissue-level remodeling in response to host immune and epithelial cell state perturbations, and bacterial population shifts, localized inflammatory responses, and metabolic process alterations during colitis. MicroCart enables a deep investigation of the intricate interplay between the host tissue and its microbiome with spatial multiomics.
Zhu B, et al., bioRxiv March 2024
doi: https://doi.org/10.1101/2024.03.04.583400
Multiplex protein imaging in tumour biology<span class='publication-meta'>de Souza, <i>et al.</i>, <i>Nat. Rev. Cancer</i> February 2024</span>
Tissue imaging has become much more colourful in the past decade. Advances in both experimental and analytical methods now make it possible to image protein markers in tissue samples in high multiplex. The ability to routinely image 40–50 markers simultaneously, at single-cell or subcellular resolution, has opened up new vistas in the study of tumour biology. Cellular phenotypes, interaction, communication and spatial organization have become amenable to molecular-level analysis, and application to patient cohorts has identified clinically relevant cellular and tissue features in several cancer types. Here, we review the use of multiplex protein imaging methods to study tumour biology, discuss ongoing attempts to combine these approaches with other forms of spatial omics, and highlight challenges in the field.
de Souza, et al., Nat. Rev. Cancer February 2024
doi: https://doi.org/10.1038/s41568-023-00657-4
Neoadjuvant CD40 Agonism Remodels the Tumor Immune Microenvironment in Locally Advanced Esophageal/Gastroesophageal Junction Cancer<span class='publication-meta'>Soto, <i>et al.</i>, <i>Cancer Res Commun.</i> January 2024</span>
Sotigalimab is an agonistic anti-CD40 mAb that can modulate antitumor immune responses. In a phase II clinical trial of sotigalimab combined with neoadjuvant chemoradiation (CRT) in locally advanced esophageal/gastroesophageal junction (E/GEJ) cancer with the primary outcome of efficacy as measured by pathologic complete response (pCR) rate, the combination induced pCR in 38% of treated patients. We investigated the mechanism of action of sotigalimab in samples obtained from this clinical trial. Tumor biopsies and peripheral blood samples were collected at baseline, following an initial dose of sotigalimab, and at the time of surgery after CRT completion from six patients. High dimensional single-cell techniques were used, including combined single-cell RNA-sequencing and proteomics (CITEseq) and multiplexed ion beam imaging, to analyze immune responses. Sotigalimab dramatically remodeled the immune compartment in the periphery and within the tumor microenvironment (TME), increasing expression of molecules related to antigen processing and presentation and altering metabolic pathways in myeloid cells. Concomitant with these changes in myeloid cells, sotigalimab treatment primed new T cell clonotypes and increased the density and activation of T cells with enhanced cytotoxic function. Sotigalimab treatment also induced a decrease in the frequency of Tregs in the TME. These findings indicate that a single dose of sotigalimab leads to enhanced antigen presentation that can activate T cells and induce new T cell clones. This restructuring of the TME provides elements which are critical to the development of effective antitumor immune responses and improved clinical outcomes.
Soto, et al., Cancer Res Commun. January 2024
doi: https://doi.org/10.1158/2767-9764.CRC-23-0550
Identification of cells of leukemic stem cell origin with non-canonical regenerative properties<span class='publication-meta'>Hollands, <i>et al.</i>, <i>Cell Reports Medicine</i> April 2024</span>
Despite most acute myeloid leukemia (AML) patients entering remission following chemotherapy, outcomes remain poor due to surviving leukemic cells that contribute to relapse. The nature of these enduring cells is poorly understood. Here, through temporal single-cell transcriptomic characterization of AML hierarchical regeneration in response to chemotherapy, we reveal a cell population: AML regeneration enriched cells (RECs). RECs are defined by CD74/CD68 expression, and although derived from leukemic stem cells (LSCs), are devoid of stem/progenitor capacity. Based on REC in situ proximity to CD34-expressing cells identified using spatial transcriptomics on AML patient bone marrow samples, RECs demonstrate the ability to augment or reduce leukemic regeneration in vivo based on transfusion or depletion, respectively. Furthermore, RECs are prognostic for patient survival as well as predictive of treatment failure in AML cohorts. Our study reveals RECs as a previously unknown functional catalyst of LSC-driven regeneration contributing to the non-canonical framework of AML regeneration.
Hollands, et al., Cell Reports Medicine April 2024
doi: https://doi.org/10.1016/j.xcrm.2024.101485
Immune profiling and tracking of two-dimensional transition metal dichalcogenides in cells and tissues <span class='publication-meta'>Gazzi, <i>et al.</i>, <i>Nano Today</i> February 2024</span>
Two-dimensional (2D) transition metal dichalcogenides (TMDs), such as molybdenum disulfide (MoS2) and tungsten disulfide (WS2), have attracted considerable interest in biomedicine due to their unique combination of physicochemical properties. The effect of nanomaterials on immune cells and their biodistribution are critical aspects of their clinical translation. However, understanding the interactions of these emerging 2D nanomaterials with the complex pool of peripheral blood mononuclear cells (PBMCs) at the single-cell level and their in situ localization in the main organs still needs to be discovered, preventing their translation in medical settings. Here, we report in-depth immune profiling of water-based and defect-free 2D formulations of MoS2 and WS2 through the simultaneous label-free tracking of their immune cell interactions, both ex vivo in human PBMCs and in vivo in mice by high-dimensional analytical approaches, as well as their biodistribution. For comparison, we studied graphene, the hitherto most explored 2D material for biomedical applications. First, we assessed the impact at the protein and gene level by multiplex protein arrays and RNA sequencing, demonstrating a very modest effect of MoS2 and WS2 on immune cell functionality compared to graphene. Then, a single-cell view of the effects of MoS2 and WS2 on 16 primary human immune cell types in terms of viability and functionality was obtained by single-cell mass cytometry by time-of-flight (CyTOF). We explored over 30 markers looking at multiple cell parameters. Finally, we present evidence that MoS2 and WS2 are visible, without the need for labeling, at the single-cell and tissue level by CyTOF, imaging mass cytometry, and multiplexed ion beam imaging by time-of-flight (MIBI-TOF). In particular, MoS2 and WS2 could be detected in the molybdenum (95Mo) and tungsten (180–186W) channels, respectively, which are not used for commercial mass cytometry tags, allowing for the simultaneous interrogation of a wide variety of biological parameters ex vivo and in vivo following intravenous administration of the TMDs. Indeed, we demonstrated the accumulation of TMDs in the main organs by MIBI-TOF and they could also be identified in specific immune cell subsets by CyTOF. Among the two TMDs studied, WS2 exhibited the highest brightness and signal intensity in all the cell subpopulations and tissues analyzed. In conclusion, we identified TMDs as immune-compatible nanoplatforms, traceable at the single-cell and tissue (sub-organ) levels, thus opening up new perspectives for their exploration in biomedicine.
Gazzi, et al., Nano Today February 2024
doi: https://doi.org/10.1016/j.nantod.2023.102084
Single-cell high-dimensional imaging mass cytometry: one step beyond in oncology<span class='publication-meta'>Glasson, <i>et al.</i>, <i>Semin Immunopathol</i> January 2023</span>
Solid tumors have a dynamic ecosystem in which malignant and non-malignant (endothelial, stromal, and immune) cell types constantly interact. Importantly, the abundance, localization, and functional orientation of each cell component within the tumor microenvironment vary significantly over time and in response to treatment. Such intratumoral heterogeneity influences the tumor course and its sensitivity to treatments. Recently, high-dimensional imaging mass cytometry (IMC) has been developed to explore the tumor ecosystem at the single-cell level. In the last years, several studies demonstrated that IMC is a powerful tool to decipher the tumor complexity. In this review, we summarize the potential of this technology and how it may be useful for cancer research (from preclinical to clinical studies).
Glasson, et al., Semin Immunopathol January 2023
doi: https://doi.org/10.1007/s00281-022-00978-w
Using random forests to uncover the predictive power of distance-varying cell interactions in tumor microenvironments<span class='publication-meta'>VanderDoes, <i>et al.</i>, <i>PLoS Comput Biol</i> June 2024</span>
Tumor microenvironments (TMEs) contain vast amounts of information on patient’s cancer through their cellular composition and the spatial distribution of tumor cells and immune cell populations. Exploring variations in TMEs between patient groups, as well as determining the extent to which this information can predict outcomes such as patient survival or treat- ment success with emerging immunotherapies, is of great interest. Moreover, in the face of a large number of cell interactions to consider, we often wish to identify specific interactions that are useful in making such predictions. We present an approach to achieve these goals based on summarizing spatial relationships in the TME using spatial K functions, and then applying functional data analysis and random forest models to both predict outcomes of interest and identify important spatial relationships. This approach is shown to be effective in simulation experiments at both identifying important spatial interactions while also control- ling the false discovery rate. We further used the proposed approach to interrogate two real data sets of Multiplexed Ion Beam Images of TMEs in triple negative breast cancer and lung cancer patients. The methods proposed are publicly available in a companion R package funkycells.
VanderDoes, et al., PLoS Comput Biol June 2024
doi: https://doi.org/10.1371/journal.pcbi.1011361
High Throughput Multi-Omics Approaches for Clinical Trial Evaluation and Drug Discovery<span class='publication-meta'>Zielinski, <i>et al.</i>, <i>Front. Immunol.</i> March 2021</span>
High throughput single cell multi-omics platforms, such as mass cytometry (cytometry by time-of-flight; CyTOF), high dimensional imaging (>6 marker; Hyperion, MIBIscope, CODEX, MACSima) and the recently evolved genomic cytometry (Citeseq or REAPseq) have enabled unprecedented insights into many biological and clinical questions, such as hematopoiesis, transplantation, cancer, and autoimmunity. In synergy with constantly adapting new single-cell analysis approaches and subsequent accumulating big data collections from these platforms, whole atlases of cell types and cellular and sub-cellular interaction networks are created. These atlases build an ideal scientific discovery environment for reference and data mining approaches, which often times reveals new cellular disease networks. In this review we will discuss how combinations and fusions of different -omic workflows on a single cell level can be used to examine cellular phenotypes, immune effector functions, and even dynamic changes, such as metabolomic state of different cells in a sample or even in a defined tissue location. We will touch on how pre- print platforms help in optimization and reproducibility of workflows, as well as community outreach. We will also shortly discuss how leveraging single cell multi-omic approaches can be used to accelerate cellular biomarker discovery during clinical trials to predict response to therapy, follow responsive cell types, and define novel druggable target pathways. Single cell proteome approaches already have changed how we explore cellular mechanism in disease and during therapy. Current challenges in the field are how we share these disruptive technologies to the scientific communities while still including new approaches, such as genomic cytometry and single cell metabolomics.
Zielinski, et al., Front. Immunol. March 2021
doi: https://doi.org/10.3389/fimmu.2021.590742
CellSighter: a neural network to classify cells in highly multiplexed images<span class='publication-meta'>Amitay, <i>et al.</i>, <i>Nat. Comm.</i> July 2023</span>
Multiplexed imaging enables measurement of multiple proteins in situ, offering an unprecedented opportunity to chart various cell types and states in tissues. However, cell classification, the task of identifying the type of individual cells, remains challenging, labor-intensive, and limiting to throughput. Here, we present CellSighter, a deep-learning based pipeline to accelerate cell classification in multiplexed images. Given a small training set of expert-labeled images, CellSighter outputs the label probabilities for all cells in new images. CellSighter achieves over 80% accuracy for major cell types across imaging platforms, which approaches inter-observer concordance. Ablation studies and simulations show that CellSighter is able to generalize its training data and learn features of protein expression levels, as well as spatial features such as subcellular expression patterns. CellSighter’s design reduces overfitting, and it can be trained with only thousands or even hundreds of labeled examples. CellSighter also outputs a prediction confidence, allowing downstream experts control over the results. Altogether, CellSighter drastically reduces hands-on time for cell classification in multiplexed images, while improving accuracy and consistency across datasets.
Amitay, et al., Nat. Comm. July 2023
doi: https://doi.org/10.1038/s41467-023-40066-7
Expanded vacuum-stable gels for multiplexed high-resolution spatial histopathology<span class='publication-meta'>Bai, <i>et al.</i>, <i>Nat. Comm.</i> July 2023</span>
Cellular organization and functions encompass multiple scales in vivo. Emerging high-plex imaging technologies are limited in resolving subcellular biomolecular features. Expansion Microscopy (ExM) and related techniques physically expand samples for enhanced spatial resolution, but are challenging to be combined with high-plex imaging technologies to enable integrative multiscaled tissue biology insights. Here, we introduce Expand and comPRESS hydrOgels (ExPRESSO), an ExM framework that allows high-plex protein staining, physical expansion, and removal of water, while retaining the lateral tissue expansion. We demonstrate ExPRESSO imaging of archival clinical tissue samples on Multiplexed Ion Beam Imaging and Imaging Mass Cytometry platforms, with detection capabilities of > 40 markers. Application of ExPRESSO on archival human lymphoid and brain tissues resolved tissue architecture at the subcellular level, particularly that of the blood-brain barrier. ExPRESSO hence provides a platform for extending the analysis compatibility of hydrogel-expanded biospecimens to mass spectrometry, with minimal modifications to protocols and instrumentation.
Bai, et al., Nat. Comm. July 2023
doi: https://doi.org/10.1038/s41467-023-39616-w
Spatial profiling technologies illuminate the tumor microenvironment<span class='publication-meta'>Elhanani, <i>et al.</i>, <i>Cancer Cell</i> March 2023</span>
The tumor microenvironment (TME) is composed of many different cellular and acellular components that together drive tumor growth, invasion, metastasis, and response to therapies. Increasing realization of the significance of the TME in cancer biology has shifted cancer research from a cancer-centric model to one that considers the TME as a whole. Recent technological advancements in spatial profiling methodologies provide a systematic view and illuminate the physical localization of the components of the TME. In this review, we provide an overview of major spatial profiling technologies. We present the types of information that can be extracted from these data and describe their applications, findings and challenges in cancer research. Finally, we provide a future perspective of how spatial profiling could be integrated into cancer research to improve patient diagnosis, prognosis, stratification to treatment and development of novel therapeutics.
Elhanani, et al., Cancer Cell March 2023
doi: https://doi.org/10.1016/j.ccell.2023.01.010
Diffuse large B-cell lymphomas have spatially defined, tumor immune microenvironments revealed by high-parameter imaging <span class='publication-meta'>Wright, <i>et al.</i>, <i>Blood Adv.</i> March 2023</span>
Diffuse large B-cell lymphoma (DLBCL) not otherwise specified is the most common aggressive non-Hodgkin lymphoma and a biologically heterogeneous disease. Despite the development of effective immunotherapies, the organization of the DLBCL tumor-immune microenvironment (TIME) remains poorly understood.We interrogated the intact TIME of 51 de novo DLBCLs with triplicate sampling to characterize 337 995 tumor and immune cells using a 27-plex antibody panel that captured cell lineage, architectural, and functional markers. We spatially assigned individual cells, identified local cell neighborhoods, and established their topographical organization in situ. We found that the organization of local tumor and immune cells can be modeled by 6 composite cell neighborhood types (CNTs). Differential CNT representation divided cases into 3 aggregate TIME categories: immune-deficient, dendritic cell–enriched (DC-enriched), and macrophage-enriched (Mac-enriched). Cases with immune-deficient TIMEs have tumor cell–rich CNTs, in which the few infiltrating immune cells are enriched near CD31+ vessels, in keeping with limited immune activity. Cases with DC-enriched TIMEs selectively include tumor cell–poor/immune cell–rich CNTs with high numbers of CD11c+ DCs and antigen-experienced T cells also enriched near CD31+ vessels, in keeping with increased immune activity. Cases with Mac-enriched TIMEs selectively include tumor cell–poor/immune cell–rich CNTs with high numbers of CD163+ macrophages and CD8 T cells throughout the microenvironment, accompanied by increased IDO-1 and LAG-3 and decreased HLA-DR expression and genetic signatures in keeping with immune evasion. Our findings reveal that the heterogenous cellular components of DLBCL are not randomly distributed but organized into CNTs that define aggregate TIMEs with distinct cellular, spatial, and functional features.
Key Points:
- DLBCLs are composed of local cell neighborhoods with distinct cellular, spatial, and functional features that define structured TIMEs.
- DLBCL cell neighborhoods structurally define immune-deficient, DC-enriched, and macrophage-enriched immune microenvironments.
Wright, et al., Blood Adv. March 2023
doi: https://doi.org/10.1182/bloodadvances.2023009813
MAPS: Pathologist-level cell type annotation from tissue images through machine learning<span class='publication-meta'>Shaban, <i>et al.</i>, <i>Biorxiv</i>, 2023</span>
Highly multiplexed protein imaging is emerging as a potent technique for analyzing protein distribution within cells and tissues in their native context. However, existing cell annotation methods utilizing high-plex spatial proteomics data are resource intensive and necessitate iterative expert input, thereby constraining their scalability and practicality for extensive datasets. We introduce MAPS (Machine learning for Analysis of Proteomics in Spatial biology), a machine learning approach facilitating rapid and precise cell type identification with human-level accuracy from spatial proteomics data. Validated on multiple in-house and publicly available MIBI and CODEX datasets, MAPS outperforms current annotation techniques in terms of speed and accuracy, achieving pathologist-level precision even for challenging cell types, including tumor cells of immune origin. By democratizing rapidly deployable and scalable machine learning annotation, MAPS holds significant potential to expedite advances in tissue biology and disease comprehension.
Shaban, et al., Biorxiv, 2023
doi: https://www.biorxiv.org/content/10.1101/2023.06.25.546474v1
Dynamic CD8+ T cell responses to cancer immunotherapy in human regional lymph nodes are disrupted in metastatic lymph nodes<span class='publication-meta'>Rahim, <i>et al.</i>, <i>Cell</i> March 2023</span>
Abstract:
CD8+ T cell responses are critical for anti-tumor immunity. While extensively profiled in the tumor microenvironment, recent studies in mice identified responses in lymph nodes (LNs) as essential; however, the role of LNs in human cancer patients remains unknown. We examined CD8+ T cells in human head and neck squamous cell carcinomas, regional LNs, and blood using mass cytometry, single-cell genomics, and multiplexed ion beam imaging. We identified progenitor exhausted CD8+ T cells (Tpex) that were abundant in uninvolved LN and clonally related to terminally exhausted cells in the tumor. After anti-PD-L1 immunotherapy, Tpex in uninvolved LNs reduced in frequency but localized near dendritic cells and proliferating intermediate-exhausted CD8+ T cells (Tex-int), consistent with activation and differentiation. LN responses coincided with increased circulating Tex-int. In metastatic LNs, these response hallmarks were impaired, with immunosuppressive cellular niches. Our results identify important roles for LNs in anti-tumor immune responses in humans.
Highlights:
- Tpex are abundant in uiLNs and clonally related to Tex-term in the TME
- After anti-PD-L1 treatment, activated Tpex and Tex-int localize near DCs in uiLNs
- uiLN anti-PD-L1 responses coincide with increased circulating Tex-int
- Tpex and Tex-int responses to anti-PD-L1 therapy are disrupted in metLNs
Rahim, et al., Cell March 2023
doi: https://doi.org/10.1016/j.cell.2023.02.021
DCIS genomic signatures define biology and correlate with clinical outcome: a Human Tumor Atlas Network (HTAN) analysis of TBCRC 038 and RAHBT cohorts<span class='publication-meta'>Strand, <i>et al.</i>, <i>Cancer Cell</i> November 2022</span>
Ductal carcinoma in situ (DCIS) is the most common precursor of invasive breast cancer (IBC), with variable propensity for progression. We perform multiscale, integrated molecular profiling of DCIS with clinical outcomes by analyzing 774 DCIS samples from 542 patients with 7.3 years median follow-up from the Translational Breast Cancer Research Consortium 038 study and the Resource of Archival Breast Tissue cohorts. We identify 812 genes associated with ipsilateral recurrence within 5 years from treatment and develop a classifier that predicts DCIS or IBC recurrence in both cohorts. Pathways associated with recurrence include proliferation, immune response, and metabolism. Distinct stromal expression patterns and immune cell compositions are identified. Our multiscale approach employed in situ methods to generate a spatially resolved atlas of breast precancers, where complementary modalities can be directly compared and correlated with conventional pathology findings, disease states, and clinical outcome.
Highlights:
- Development of a new classifier for DCIS recurrence or progression
- Four stroma-specific signatures identified
- Outcome-associated pathways identified across multiple data types and compartments
- CNAs characterize DCIS subgroups associated with high-risk invasive cancers
Strand, et al., Cancer Cell November 2022
doi: https://doi.org/10.1016/j.ccell.2022.10.021
Spatial epitope barcoding reveals subclonal tumor patch behaviors<span class='publication-meta'>Rovira-Clavé, <i>et al.</i>, <i>Cancer Cell</i> November 2022</span>
Abstract: Intratumoral heterogeneity is a seminal feature of human tumors contributing to tumor progression and response to treatment. Current technologies are still largely unsuitable to accurately track phenotypes and clonal evolution within tumors, especially in response to genetic manipulations. Here, we developed epitopes for imaging using combinatorial tagging (EpicTags), which we coupled to multiplexed ion beam imaging (EpicMIBI) for in situ tracking of barcodes within tissue microenvironments. Using EpicMIBI, we dissected the spatial component of cell lineages and phenotypes in xenograft models of small cell lung cancer. We observed emergent properties from mixed clones leading to the preferential expansion of clonal patches for both neuroendocrine and non-neuroendocrine cancer cell states in these models. In a tumor model harboring a fraction of PTEN-deficient cancer cells, we observed a non-autonomous increase of clonal patch size in PTEN wild-type cancer cells. EpicMIBI facilitates in situ interrogation of cell-intrinsic and cell-extrinsic processes involved in intratumoral heterogeneity.
Rovira-Clavé, et al., Cancer Cell November 2022
doi: https://doi.org/10.1016/j.ccell.2022.09.014
Single-cell spatial proteomic imaging for human neuropathology <span class='publication-meta'>Vijayaragavan & Cannon, <i>et al.</i>, <i>Acta Neuropathol Commun</i> November 2022</span>
Abstract: Neurodegenerative disorders are characterized by phenotypic changes and hallmark proteopathies. Quantifying these in archival human brain tissues remains indispensable for validating animal models and understanding disease mechanisms. We present a framework for nanometer-scale, spatial proteomics with multiplex ion beam imaging (MIBI) for capturing neuropathological features. MIBI facilitated simultaneous, quantitative imaging of 36 proteins on archival human hippocampus from individuals spanning cognitively normal to dementia. Customized analysis strategies identified cell types and proteopathies in the hippocampus across stages of Alzheimer’s disease (AD) neuropathologic change. We show microglia-pathologic tau interactions in hippocampal CA1 subfield in AD dementia. Data driven, sample independent creation of spatial proteomic regions identified persistent neurons in pathologic tau neighborhoods expressing mitochondrial protein MFN2, regardless of cognitive status, suggesting a survival advantage. Our study revealed unique insights from multiplexed imaging and data-driven approaches for neuropathologic analysis and serves broadly as a methodology for spatial proteomic analysis of archival human neuropathology.
Vijayaragavan, K. & Cannon, B.J., et al., Acta Neuropathologica Communications Nov 2022
doi: https://www.ionpath.com/wp-content/uploads/2023/01/Ionpath-MIBI-Oveview-221109.pdf
Single-nucleus and spatial transcriptomics of archival pancreatic cancer reveals multi-compartment reprogramming after neoadjuvant treatment<span class='publication-meta'>Hwang, <i>et al., Nature Genetics</i> July 2022</span>
Hwang, et al., Nature Genetics July 2022
doi: https://doi.org/10.1038/s41588-022-01134-8
An approach that combines protein and nucleic acid imaging reveals virus-dependent B cell and macrophage immunosuppression of tissue microenvironments <span class='publication-meta'>Jiang, <i>et al.</i>, <i>Immunity</i> April 2022</span>
Abstract: Understanding the mechanisms of HIV tissue persistence necessitates the ability to visualize tissue microenvironments where infected cells reside; however, technological barriers limit our ability to dissect the cellular components of these HIV reservoirs. Here, we developed protein and nucleic acid in situ imaging (PANINI) to simultaneously quantify DNA, RNA, and protein levels within these tissue compartments. By coupling PANINI with multiplexed ion beam imaging (MIBI), we measured over 30 parameters simultaneously across archival lymphoid tissues from healthy or simian immunodeficiency virus (SIV)-infected nonhuman primates. PANINI enabled the spatial dissection of cellular phenotypes, functional markers, and viral events resulting from infection. SIV infection induced IL-10 expression in lymphoid B cells, which correlated with local macrophage M2 polarization. This highlights a potential viral mechanism for conditioning an immunosuppressive tissue environment for virion production. The spatial multimodal framework here can be extended to decipher tissue responses in other infectious diseases and tumor biology.
Jiang, et al., Immunity April 2022
doi: https://doi.org/10.1016/j.immuni.2022.03.020
Reproducible, high-dimensional imaging in archival human tissue by Multiplexed Ion Beam Imaging by Time-of-Flight (MIBI-TOF) <span class='publication-meta'>Liu, <i>et al.</i>, <i>Lab. Invest.</i> March 2022</span>
Abstract: Multiplexed ion beam imaging by time-of-flight (MIBI-TOF) is a form of mass spectrometry imaging that uses metal labeled antibodies and secondary ion mass spectrometry to image dozens of proteins simultaneously in the same tissue section. Working with the National Cancer Institute’s (NCI) Cancer Immune Monitoring and Analysis Centers (CIMAC), we undertook a validation study, assessing concordance across a dozen serial sections of a tissue microarray of 21 samples that were independently processed and imaged by MIBI-TOF or single-plex immunohistochemistry (IHC) over 12 days. Pixel-level features were highly concordant across all 16 targets assessed in both staining intensity (R2 = 0.94 ± 0.04) and frequency (R2 = 0.95 ± 0.04). Comparison to digitized, single-plex IHC on adjacent serial sections revealed similar concordance (R2 = 0.85 ± 0.08) as well. Lastly, automated segmentation and clustering of eight cell populations found that cell frequencies between replicates yielded an average correlation of R2 = 0.92 ± 0.06. Taken together, we demonstrate that MIBI-TOF, with well-vetted reagents and automated analysis, can generate consistent and quantitative annotations of clinically relevant cell states in archival human tissue, and more broadly, present a scalable framework for benchmarking multiplexed IHC approaches.
Liu, et al., Lab. Invest. March 2022
doi: https://doi.org/10.1038/s41374-022-00778-8
The immunoregulatory landscape of human tuberculosis granulomas <span class='publication-meta'>McCaffrey, <i>et al.</i>, <i>Nature Immunology</i> February 2022</span>
Abstract: Tuberculosis (TB) in humans is characterized by formation of immune-rich granulomas in infected tissues, the architecture and composition of which are thought to affect disease outcome. However, our understanding of the spatial relationships that control human granulomas is limited. Here, we used multiplexed ion beam imaging by time of flight (MIBI-TOF) to image 37 proteins in tissues from patients with active TB. We constructed a comprehensive atlas that maps 19 cell subsets across 8 spatial microenvironments. This atlas shows an IFN-γ-depleted microenvironment enriched for TGF-β, regulatory T cells and IDO1+ PD-L1+ myeloid cells. In a further transcriptomic meta-analysis of peripheral blood from patients with TB, immunoregulatory trends mirror those identified by granuloma imaging. Notably, PD-L1 expression is associated with progression to active TB and treatment response. These data indicate that in TB granulomas, there are local spatially coordinated immunoregulatory programs with systemic manifestations that define active TB.
McCaffrey, et al., Nature Immunology February 2022
doi: https://doi.org/10.1038/s41590-021-01121-x
Transition to invasive breast cancer is associated with progressive changes in the structure and composition of tumor stroma <span class='publication-meta'>Risom, <i>et al.</i>, <i>Cell</i> January 2022</span>
Abstract: Ductal carcinoma in situ (DCIS) is a pre-invasive lesion that is thought to be a precursor to invasive breast cancer (IBC). To understand the changes in the tumor microenvironment (TME) accompanying transition to IBC, we used multiplexed ion beam imaging by time of flight (MIBI-TOF) and a 37-plex antibody staining panel to interrogate 79 clinically annotated surgical resections using machine learning tools for cell segmentation, pixel-based clustering, and object morphometrics. Comparison of normal breast with patientmatched DCIS and IBC revealed coordinated transitions between four TME states that were delineated based on the location and function of myoepithelium, fibroblasts, and immune cells. Surprisingly, myoepithelial disruption was more advanced in DCIS patients that did not develop IBC, suggesting this process could be protective against recurrence. Taken together, this HTAN Breast PreCancer Atlas study offers insight into drivers of IBC relapse and emphasizes the importance of the TME in regulating these processes.
Risom, et al., Cell January 2022
doi: https://doi.org/10.1016/j.cell.2021.12.023
Multiplexed Ion Beam Imaging: Insights into Pathobiology <span class='publication-meta'>Liu, <i>et al.</i>, <i>Annual Review of Pathology</i> 2022</span>
Abstract: Next-generation tools formultiplexed imaging have driven a new wave of innovation in understanding how single-cell function and tissue structure are interrelated. In previous work, we developed multiplexed ion beam imaging by time of flight, a highly multiplexed platform that uses secondary ion mass spectrometry to image dozens of antibodies tagged with metal reporters. As instrument throughput has increased, the breadth and depth of imaging data have increased as well. To extract meaningful information from these data, we have developed tools for cell identification, cell classification, and spatial analysis. In this review, we discuss these tools and provide examples of their application in various contexts, including ductal carcinoma in situ, tuberculosis, and Alzheimer’s disease.We hope the synergy between multiplexed imaging and automated image analysis will drive a new era in anatomic pathology and personalized medicine wherein quantitative spatial signatures are used routinely for more accurate diagnosis, prognosis, and therapeutic selection.
Liu, et al., Annual Review of Pathology 2022
doi: https://doi.org/10.1146/annurev-pathmechdis-030321-091459
Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer<span class='publication-meta'>Chan, <i>et al.</i>, <i>Cancer Cell</i> October 2021</span>
Abstract: Small cell lung cancer (SCLC) is an aggressive malignancy that includes subtypes defined by differential expression of ASCL1, NEUROD1, and POU2F3 (SCLC-A, -N, and -P, respectively). To define the heterogeneity of tumors and their associated microenvironments across subtypes, we sequenced 155,098 transcriptomes from 21 human biospecimens, including 54,523 SCLC transcriptomes. We observe greater tumor diversity in SCLC than lung adenocarcinoma, driven by canonical, intermediate, and admixed subtypes. We discover a PLCG2-high SCLC phenotype with stem-like, pro-metastatic features that recurs across subtypes and predicts worse overall survival. SCLC exhibits greater immune sequestration and less immune infiltration than lung adenocarcinoma, and SCLC-N shows less immune infiltrate and greater T cell dysfunction than SCLC-A. We identify a profibrotic, immunosuppressive monocyte/macrophage population in SCLC tumors that is particularly associated with the recurrent, PLCG2-high subpopulation.
Chan, et al., Cancer Cell October 2021
doi: https://doi.org/10.1016/j.ccell.2021.09.008
Multiplexed imaging reveals an IFN-γ-driven inflammatory state in nivolumab-associated gastritis<span class='publication-meta'>Ferrian, <i>et al.</i>, <i>Cell Reports Medicine</i> October 2021</span>
Abstract: Immune checkpoint blockade using PD-1 inhibition is an effective approach for treating a wide variety of cancer subtypes. While lower gastrointestinal (GI) side effects are more common, upper gastrointestinal adverse events are rarely reported. Here, we present a case of nivolumab-associated autoimmune gastritis. To elucidate the immunology underlying this condition, we leverage multiplexed ion beam imaging by time-of-flight (MIBI-TOF) to identify the presence and proportion of infiltrating immune cells from a single section of biopsy specimen. Using MIBI-TOF, we analyze formalin-fixed, paraffin-embedded human gastric tissue with 28 labels simultaneously. Our analyses reveal a gastritis characterized by severe mucosal injury, interferon gamma (IFN-γ)-producing gastric epithelial cells, and mixed inflammation that includes CD8 and CD4 T cell infiltrates with reduced expression of granzyme B and FOXP3, respectively. Here, we provide a comprehensive multiplexed histopathological mapping of gastric tissue, which identifies IFN-γ-producing epithelial cells as possible contributors to the nivolumab-associated gastritis.
Ferrian, et al., Cell Reports Medicine October 2021
doi: https://doi.org/10.1016/j.xcrm.2021.100419
Spatio-temporal coordination at the maternal-fetal interface promotes trophoblast invasion and vascular remodeling in the first half of human pregnancy<span class='publication-meta'>Greenbaum, <i>et al.</i>, <i>BioRxiv</i> September 2021</span>
Abstract:Beginning in the first trimester, fetally derived extravillous trophoblasts (EVTs) invade the uterus and remodel its spiral arteries, transforming them into large, dilated blood vessels that lack smooth muscle and are partially lined with EVTs instead of vascular endothelium. Several mechanisms have been proposed to explain how EVTs coordinate with decidual cells to promote a tissue microenvironment conducive to spiral artery remodeling (SAR). However, it remains a matter of debate which immune and stromal cell types participate in these interactions, how this process evolves with respect to gestational age, and which anatomic routes are the predominate path of EVT invasion in humans. To elucidate this complex interplay, we used multiplexed ion beam imaging by time of flight with a 37-plex antibody panel to build the first spatio-temporal atlas of the human maternal-fetal interface in the first half of pregnancy at single-cell resolution. We analyzed ~500,000 cells and 588 spiral arteries within intact decidua from 66 patients between 6-20 weeks of gestation. Using custom machine learning algorithms for cell segmentation and classification, we evaluated the spatial distributions and phenotype of 20 maternal and five EVT populations with respect to gestational age and SAR. Gestational age substantially influenced the frequency of most maternal immune and stromal cells, with tolerogenic subsets expressing CD206, CD163, TIM-3, Galectin-9, and IDO-1 preferentially enriched at later time points. In contrast, SAR progression, and not gestational age, preferentially correlated with local invasion of EVTs. Lastly, by comparing spatial co-occurrence and phenotype of decidual interstitial, perivascular and intravascular EVTs with respect to SAR progression, we developed a statistical model suggesting an intravasation mechanism as the predominant route of EVT invasion in superficial decidua. Taken together, these results support a coordinated model of decidualization in which increasing gestational age drives a transition in maternal decidua towards a tolerogenic niche conducive to locally regulated, EVT-dependent SAR.
Greenbaum, et al., BioRxiv September 2021
doi: https://doi.org/10.1101/2021.09.08.459490
Subcellular localization of biomolecules and drug distribution by high-definition ion beam imaging<span class='publication-meta'>Rovira-Clavé, <i>et al.</i>, <i>Nature Communications</i> July 2021</span>
Abstract: Simultaneous visualization of the relationship between multiple biomolecules and their ligands or small molecules at the nanometer scale in cells will enable greater understanding of how biological processes operate. We present here high-definition multiplex ion beam imaging (HD-MIBI), a secondary ion mass spectrometry approach capable of high-parameter imaging in 3D of targeted biological entities and exogenously added structurally-unmodified small molecules. With this technology, the atomic constituents of the biomolecules themselves can be used in our system as the “tag” and we demonstrate measurements down to ~30 nm lateral resolution. We correlated the subcellular localization of the chemotherapy drug cisplatin simultaneously with five subnuclear structures. Cisplatin was preferentially enriched in nuclear speckles and excluded from closed-chromatin regions, indicative of a role for cisplatin in active regions of chromatin. Unexpectedly, cells surviving multi-drug treatment with cisplatin and the BET inhibitor JQ1 demonstrated near total cisplatin exclusion from the nucleus, suggesting that selective subcellular drug relocalization may modulate resistance to this important chemotherapeutic treatment. Multiplexed high-resolution imaging techniques, such as HD-MIBI, will enable studies of biomolecules and drug distributions in biologically relevant subcellular microenvironments by visualizing the processes themselves in concert, rather than inferring mechanism through surrogate analyses.
Rovira-Clavé, et al., Nature Communications July 2021
doi: https://doi.org/10.1038/s41467-021-24822-1
On Clustering for Cell Phenotyping in Multiplex Immunohistochemistry (mIHC) and Multiplexed Ion Beam Imaging (MIBI) Data<span class='publication-meta'>Seal, <i>et al.</i>, <i>Research Square</i> July 2021</span>
Results: Unsupervised cell phenotyping methods including PhenoGraph, flowMeans, and SamSPECTRAL, primarily used in flow cytometry data, often perform poorly or need elaborate tuning to perform well in the context of mIHC and MIBI data. We show that, instead, semi-supervised cell clustering using Random Forests, linear and quadratic discriminant analysis are superior. We test the performance of the methods on two mIHC datasets from the University of Colorado School of Medicine and a publicly available MIBI dataset. Each dataset contains numerous highly complex images.
Seal, et al., Research Square July 2021
doi: https://doi.org/10.21203/rs.3.rs-609920/v1
Multiplexed single-cell metabolic profiles organize the spectrum of cytotoxic human T cells<span class='publication-meta'>Hartmann, <i>et al., Nat. Biotechnology</i> Aug 2020</span>
Hartmann, et al., Nat. Biotechnol. 31 Aug 2020: s41587-020-0651-8
doi: 10.1038/s41587-020-0651-8
Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma<span class='publication-meta'>Ji, <i>et al., Cell</i> Jul 2020</span>
Ji, et al., Cell (2020) July 23
Multiplexed imaging of human tuberculosis granulomas uncovers immunoregulatory features conserved across tissue and blood<span class='publication-meta'>McCaffrey, <i>et al., BioRxiv</i> Jun 2020</span>
PREPRINT – McCaffrey, et al., BioRxiv (2020) June 09
MIBI-TOF: A multiplexed imaging platform relates cellular phenotypes and tissue structure<span class='publication-meta'>Keren, <i>et al., Science Advances</i> Oct 2019</span>
Abstract: Understanding tissue structure and function requires tools that quantify the expression of multiple proteins while preserving spatial information. Here, we describe MIBI-TOF (multiplexed ion beam imaging by time of flight), an instrument that uses bright ion sources and orthogonal time-of-flight mass spectrometry to image metal-tagged antibodies at subcellular resolution in clinical tissue sections. We demonstrate quantitative, full periodic table coverage across a five-log dynamic range, imaging 36 labeled antibodies simultaneously with histochemical stains and endogenous elements. We image fields of view up to 800 μm × 800 μm at resolutions down to 260 nm with sensitivities approaching single-molecule detection. We leverage these properties to interrogate intrapatient heterogeneity in tumor organization in triple-negative breast cancer, revealing regional variability in tumor cell phenotypes in contrast to a structured immune response. Given its versatility and sample back-compatibility, MIBI-TOF is positioned to leverage existing annotated, archival tissue cohorts to explore emerging questions in cancer, immunology, and neurobiology.
Keren, et al., Science Advances 09 Oct 2019: 5(10), eaax5851
Tissue imaging by mass spectrometry: a practical guide for the medicinal chemist<span class='publication-meta'>Johnson, <em>et al., ACS Medical Chemistry Letters</em> Jan 2019</span>
Johnson et al. ACS Medical Chemistry Letters. January, 2019
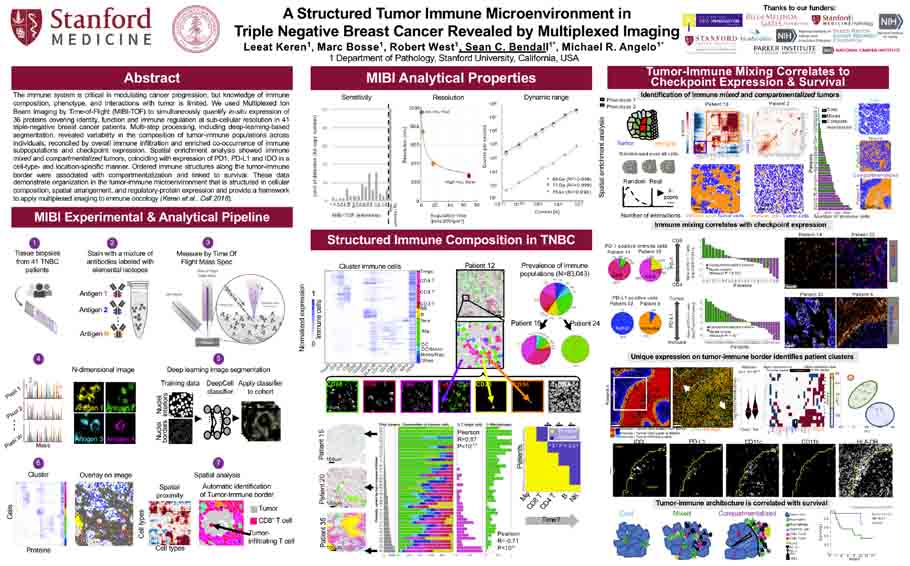
A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging<span class='publication-meta'>Keren, <em>et al, Cell</em> Sept 2018</span>
Keren et al. Cell, Volume 174, Issue 6, P1373-1387. E19. September 06, 2018
Multiplexed ion beam imaging analysis for quantitation of protein expression in cancer tissue sections<span class='publication-meta'>Rost, <em>et al., Lab. Invest.</em> Aug 2017</span>
Rost et al., Lab Invest. 2017 Aug;97(8):992-1003
SITC 2022 - #89
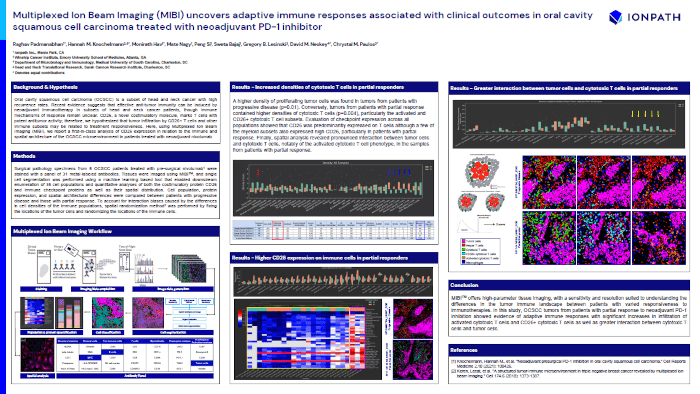
Multiplexed Ion Beam Imaging (MIBI) uncovers adaptive immune responses associated with clinical outcomes in oral cavity squamous cell carcinoma treated with neoadjuvant PD-1 inhibitor
SITC 2022 - #1294
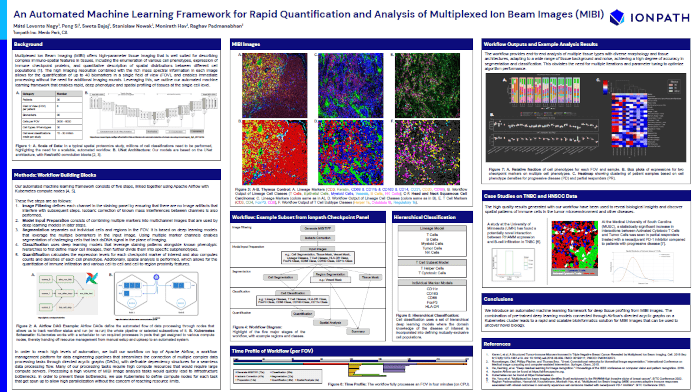
An Automated Machine Learning Framework for Rapid Quantification and Analysis of Multiplexed Ion Beam Images (MIBI)
SITC 2021
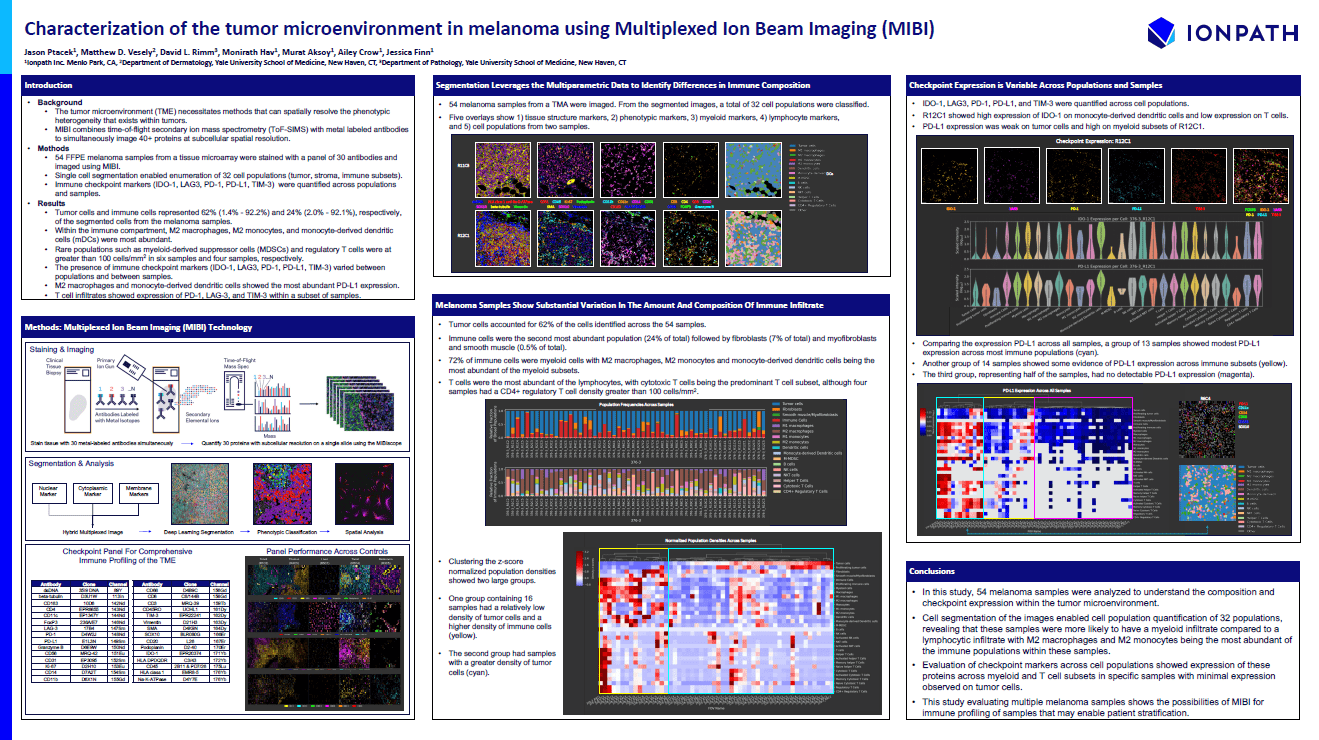
Characterization of the Tumor Microenvironment in Melanoma Using Multiplexed Ion Beam Imaging (MIBI)
SITC 2020 - P48
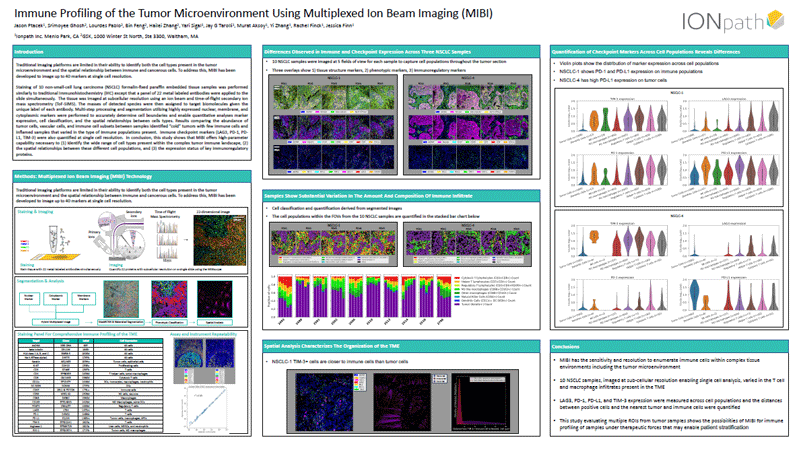
Advances in Multiplexed Ion Beam Imaging (MIBI) for immune profiling of the tumor microenvironment
SITC 2019 - P61
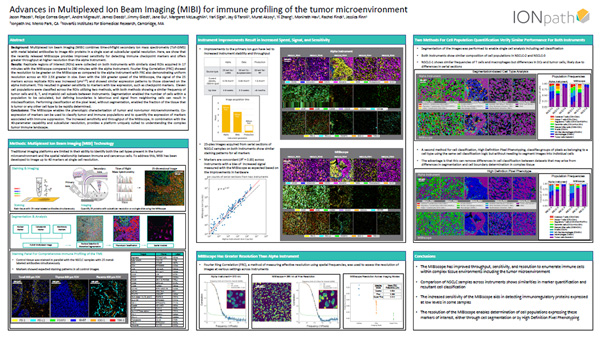
Segmentation and Classification of Single Cells using Multiplexed Ion Beam Imaging (MIBI)
SITC 2018 - P106
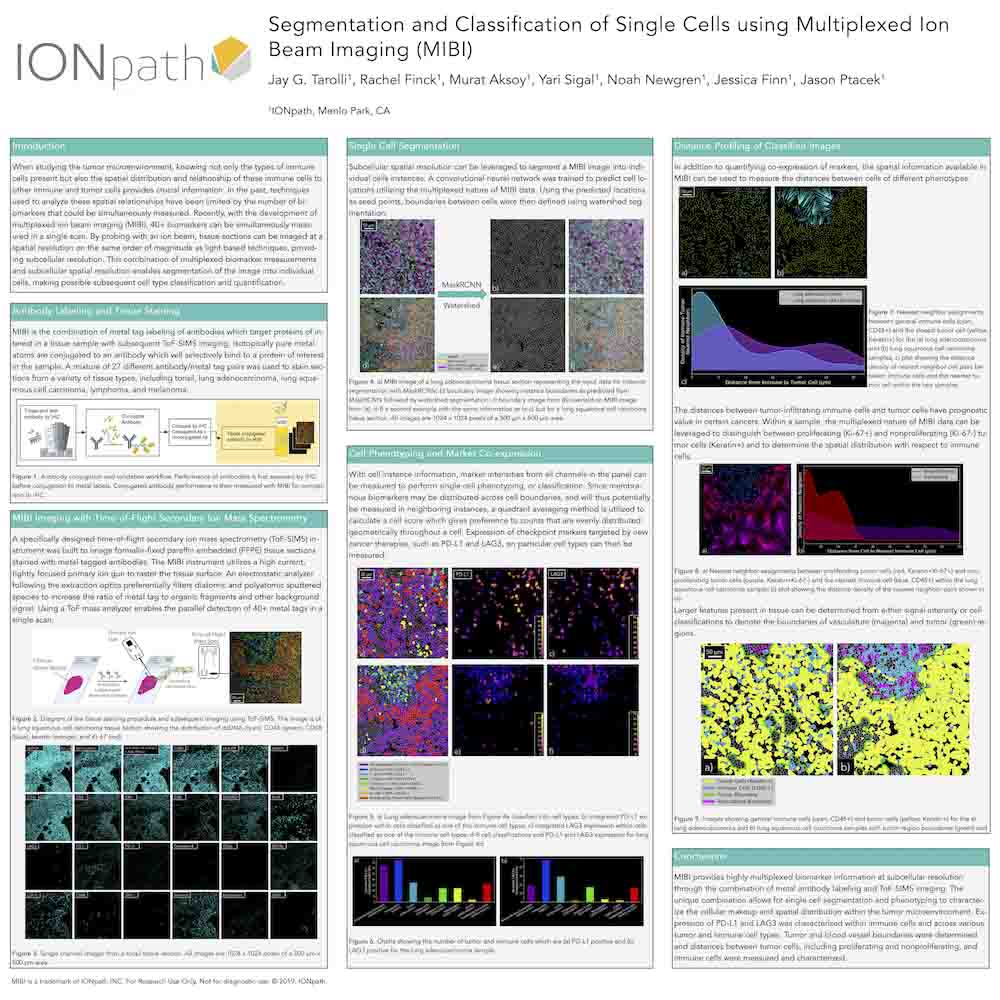
Multiplexed Ion Beam Imaging For Characterization of the Tumor Microenvironment Across Tumor Types
SITC 2018 - P20
A Structured Tumor Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Imaging