New Cancer Atlas Illuminates Small Cell Lung Cancer’s Spread with MIBI Technology
A new molecular map of small cell lung cancer (SCLC) is shedding light on how the aggressive malignancy spreads so quickly and why it’s so difficult to treat. Employing single-cell transcriptome sequencing and multiplexed ion beam imaging (MIBI™) high-definition spatial proteomic technology, an international group of researchers led by Memorial Sloan Kettering Cancer Center has discovered that an abundance of rare stem-like cells within SCLC tumors is linked to a poor prognosis. Only a tiny fraction of these cells may be needed to drive metastatic behavior across tumors.
Published in Cancer Cell, “Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer,” is a recent paper out of the Human Tumor Atlas Network Consortium. This group of cancer centers from across the United States is working to develop high-resolution maps of cancer tumor terrains — providing a clearer picture of how cancers change over time to become more deadly.
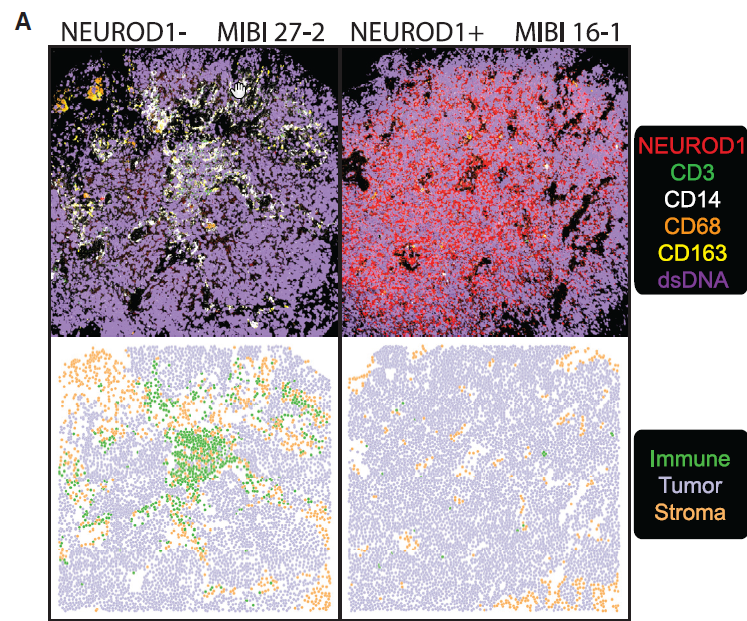
Complex heterogeneity observed and a common PLCG2+ cluster was discovered
The picture of SCLC that emerged from the group’s work is one of surprisingly complex heterogeneity and marked metastasis. In order to define the heterogeneity of tumors and their associated microenvironments across subtypes, the researchers sequenced 155,098 transcriptomes from 21 human biospecimens, including 54,523 SCLC transcriptomes. They found that the top differentially upregulated gene within a particularly aggressive cluster common to many samples, phospholipase C gamma 2 (PLCG2), is associated with increased stem-like and pro-metastatic potential.
To determine the clinical significance of PLCG2 expression, the scientists used MIBI imaging on a tissue microarray representing an independent cohort of SCLC tumor specimens. Results indicated that a PLCG2-high recurrent population enlists diverse gene programs to define pronounced heterogeneity and facilitate metastasis in a profoundly immunosuppressed tumor microenvironment. In other words, these cells have molecular characteristics that make them more likely to spread.
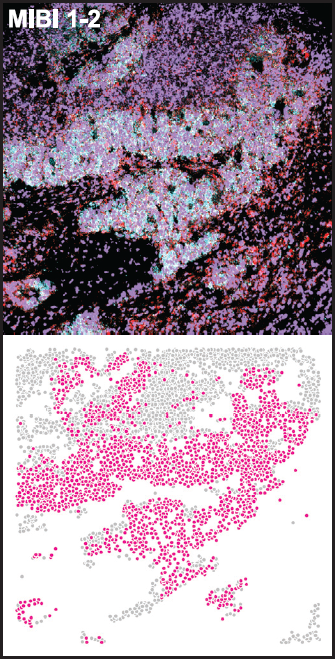
Potentially universal characteristics of malignancy revealed using MIBI
They also identified and validated the association of PLCG2-positive SCLC cells by applying MIBI to an independent cohort of SCLC tumors, differentiating the populations by specific immune cell types.
“Despite substantial clinical heterogeneity in patients with SCLC, we detected a subpopulation that was shared among tumors across subtypes, treatments, and tissue locations, pointing to a potentially universal characteristic of this malignancy,” note the researchers. “Our dataset has potential implications for the design of novel targeted therapies and immunotherapeutic approaches.”
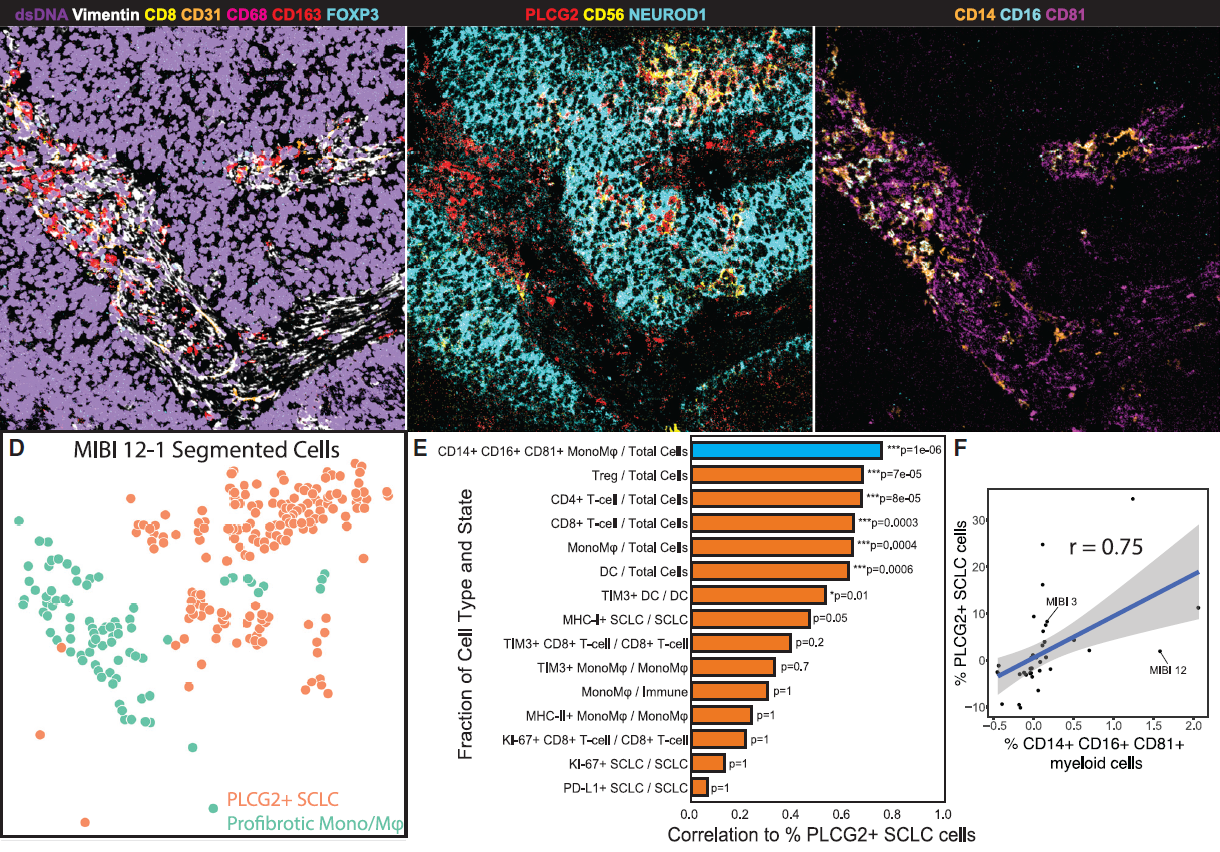
The future role of spatial proteomics using MIBI technology
By profiling multiple protein markers simultaneously at single-cell resolution in the spatial context of tissue, MIBI played an important role in the discoveries and proved to be a significant complement to scRNAseq technology.
This work could help pave the way for the development of better targeted therapies and interventions for SCLC patients. It’s a critical need, since two-thirds of cases are already metastatic at the time of diagnosis and traditional chemotherapy improves median survival only modestly. This pioneering atlas shows how large-scale studies — along with MIBI technology for spatial proteomics — could make a real difference.